
NEXTFLEX RNA-seq 2.0 UDI barcodes (8NT index, 97-192)



The NEXTFLEX™ RNA-Seq 2.0 UDI Barcodes are indexed adapters for single or paired-end sequencing with the NEXTFLEX RNA-Seq 2.0 workflow. Each 8-nt index pair is separated by at least a three-base edit distance to enable error-tolerant demultiplexing and to limit index hopping on patterned flow cells. The adapters are compatible TruSeq-style T-overhang workflows and have been qualified on all Illumina® instruments as well as Element Biosciences’ AVITI™ system. Across the full line of adapters, up to 384 unique indices are offered. Every lot is sequence-verified for index purity to protect data integrity.
| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | Barcodes |
The NEXTFLEX™ RNA-Seq 2.0 UDI Barcodes are indexed adapters for single or paired-end sequencing with the NEXTFLEX RNA-Seq 2.0 workflow. Each 8-nt index pair is separated by at least a three-base edit distance to enable error-tolerant demultiplexing and to limit index hopping on patterned flow cells. The adapters are compatible TruSeq-style T-overhang workflows and have been qualified on all Illumina® instruments as well as Element Biosciences’ AVITI™ system. Across the full line of adapters, up to 384 unique indices are offered. Every lot is sequence-verified for index purity to protect data integrity.



Product information
Overview
- UDI full-length adapters for multiplexing up to 384 libraries on Illumina® and Element® AVITI™ platforms
- Tested with the NEXTFLEX RNA-Seq 2.0 kit and compatible with TruSeq-style T-overhang library preps
- Unique dual-index design with ≥3-base edit distance reduces index hopping and sample mis-assignment on patterned flow cells
- Each lot is sequence-verified for index purity by NGS-based QC
- Compatible with NEXTFLEX Universal Blockers
The NEXTFLEX® RNA-seq 2.0 UDI barcodes (8-nt index) are conveniently plated at 6.25 µM stock concentration to be used with the NEXTFLEX® Rapid Directional RNA-Seq kit 2.0 and the CRISPRclean® Plus Stranded Total RNA Prep with rRNA Depletion.
Additional product information
Error-resistant indexing supports cleaner RNA-seq data
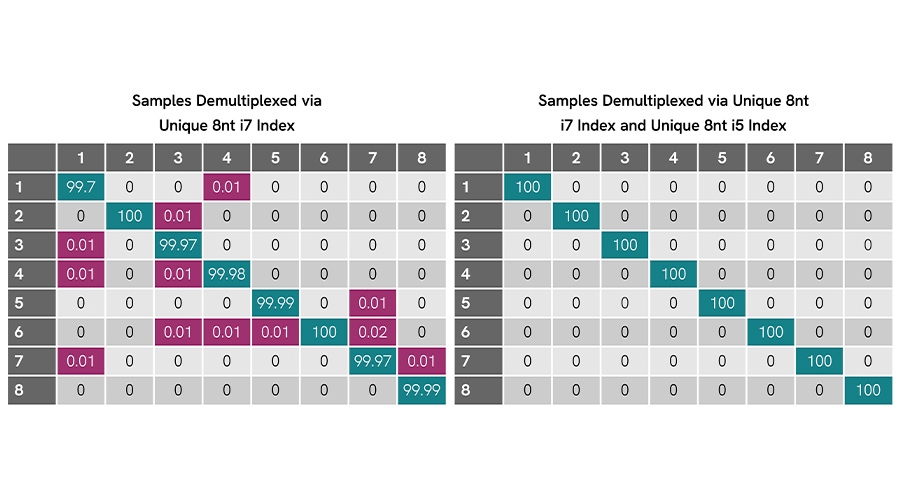
The NEXTFLEX RNA-Seq 2.0 UDI Barcodes are built on a dual 8-nt index architecture, with each pair separated by a Hamming distance of three or more bases. This spacing helps prevent single-base substitution errors from producing false index assignments during demultiplexing. In addition, requiring matched i7 and i5 index pairs ensures that reads are only assigned when both ends correspond to the correct sample. This dramatically reduces the risk of misassignment from index hopping or low-level contamination, issues that are especially problematic on patterned flow cells.

Figure 1. Dual-index design eliminates misassignment on Illumina® MiSeq®. Eight libraries prepared with NEXTFLEX Barcodes show low off-target assignment (≤0.02%) when demultiplexed using i7-only indices (left). When demultiplexed with matched i7 + i5 index pairs (right), all reads are correctly assigned with no detectable cross-contamination.
These design features contribute directly to data quality. Researchers retain more usable reads, avoid index collisions, and maintain better sample balance across RNA-seq libraries. The result is greater confidence in read-level assignments for applications such as differential expression, isoform detection, and low-input RNA profiling, where even low rates of cross-sample contamination can compromise biological interpretation.
Seamless workflow integration
NEXTFLEX RNA-Seq 2.0 UDI Barcodes are compatible with any TruSeq-style T-overhang ligation workflow, including PCR-free and amplified RNA-seq protocols. The plate-based format supports flexible library prep planning, whether you're running high-throughput batches or low-plex experiments, while preserving index integrity across stored wells.
Quality-controlled barcodes for trusted results
Each index set undergoes sequencing-based quality control to confirm purity and assignment accuracy. Proprietary contamination reduction methods lower cross-sample crosstalk to ~ 0.1%. Color balancing and avoidance of long homopolymers further support even signal distribution and reduce sequencing noise. Controlled manufacturing ensures traceable, lot-to-lot consistency for reproducible results across runs.
Scalability, blocker compatibility, and UMI options
NEXTFLEX RNA-Seq 2.0 indexing scales from 24 or 96 UDI pairs up to 384 unique combinations across four plates, without changing demultiplexing settings or workflow parameters. For added hybridization specificity, NEXTFLEX Universal Blockers can be used to improve on-target rates. For experiments requiring molecule-level quantitation, NEXTFLEX UDI-UMI Adapters incorporate UMIs directly into the indexing structure to support duplicate collapsing and accurate counting.
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Barcodes |
97 - 192
|
| Product Group |
Barcodes
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Unit Size |
192 rxns
|
Citations
- Bond, D. M., Ortega-Recalde, O., Laird, M. K., Richardson, K. S., Reese, F. C. B., Kyle, B., … Hore, T. A. (2023). The admixed brushtail possum genome reveals invasion history in New Zealand and novel imprinted genes. Nature Communications, 14, 6364. https://doi.org/10.1038/s41467-023-41784-8
- Herneisen, A. L., Li, Z.-H., Chan, A. W., Moreno, S. N. J., & Lourido, S. (2024). SPARK regulates AGC kinases central to the Toxoplasma gondii asexual cycle. eLife, 13, RP93877. https://doi.org/10.7554/eLife.93877
- Wood, J. C., et al. (2025). Chromosome-scale genome assembly for yellow wood sorrel (Oxalis stricta L.). bioRxiv. https://doi.org/10.1101/2025.06.26.661616
FAQs
-
How many unique dual index pairs are available for the RNA-Seq 2.0 UDI sets?
-
What is the index length and spacing, and why does it matter?
-
Which sequencers and software are these indices compatible with?
-
Can these barcodes be used as a drop-in with TruSeq-style T-overhang workflows?
-
Do you verify index purity?
Resources
Are you looking for resources, click on the resource type to explore further.
This excel file includes the sequences of the indexes for the 384 NEXTFLEX UDI Barcodes.


Loading...
How can we help you?
We are here to answer your questions.






























