
Gel-free library prep using miRNA isolated from exosomes
Extracellular vesicles are membrane-bound compartments which are being increasingly recognized as important players in cell-to-cell communication. They are secreted by most cell types and are present in many and perhaps all body fluids, including plasma. Extracellular vesicles are classified in different types according to their size.
Key takeaways:
- Gel-free workflow from RNA isolated from exosomes
- Exceptional miRNA discovery
To view the full content please answer a few questions
Introduction
The most characterized are exosomes, which are 50-200 nm in size. Exosomes transport a cargo of different proteins and RNA (exoRNA), mainly mRNA and miRNA. miRNAs in particular are being studied as potential biomarkers for diseases. The fact that they are present in exosomes make them attractive candidates as biomarker molecule as this means that can be collected easily and in a minimally invasive way1-3.
There is a need of a convenient method to construct libraries from exoRNA. The amount of RNA obtained from exosomes is low, with just a few picograms corresponding to miRNA. Based on quantitative analysis this can be as little as 1 miRNA molecule/100 exosomes4, making this a very challenging sample type.
Here, we describe a simplified, commercially available protocol encompassing exoRNA extraction and preparation of exosomal miRNA-seq libraries from serum and bone marrow.
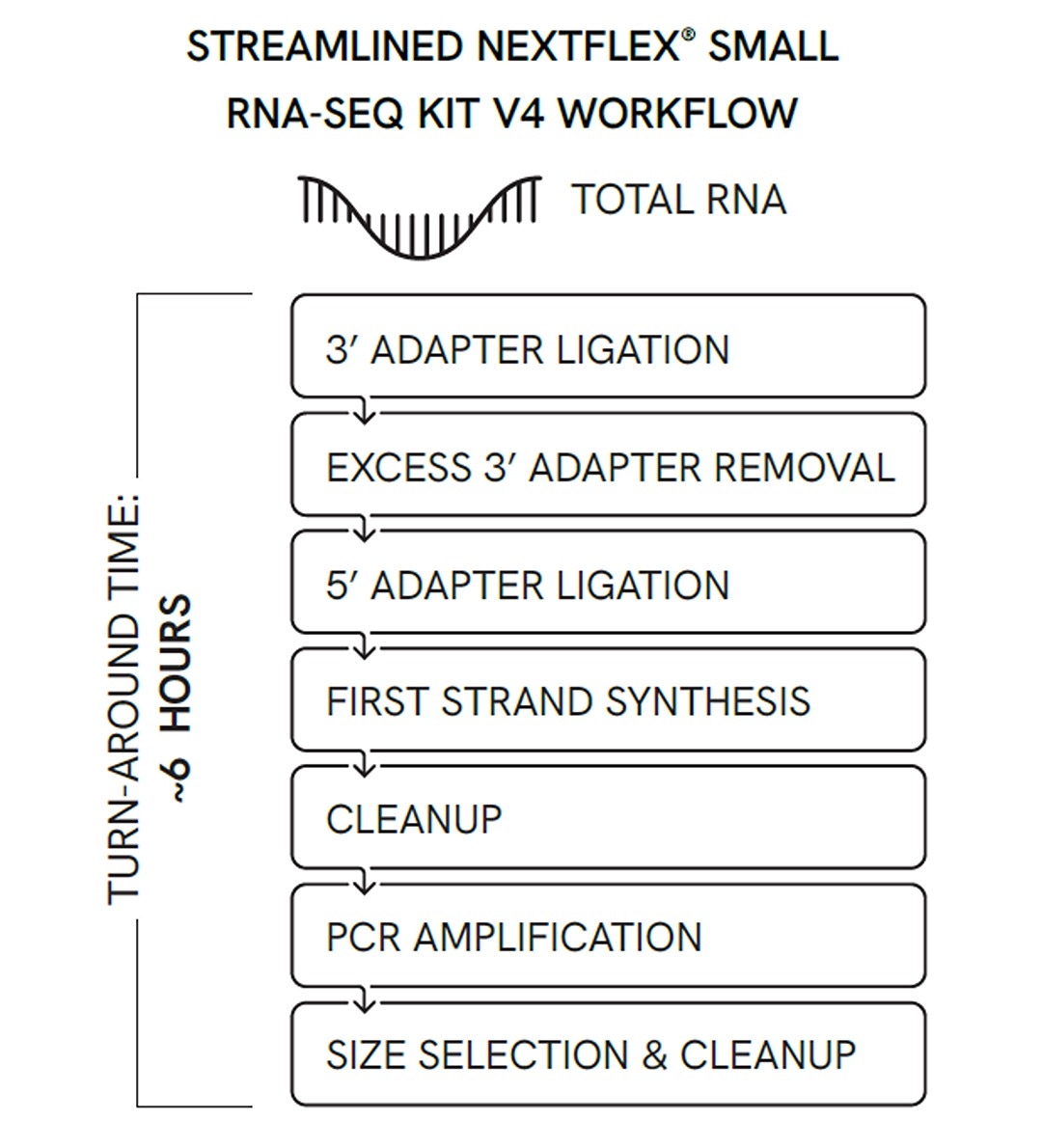
The NEXTFLEX® Small RNA-Seq Kit v4
 NEXTFLEX Small RNA Sequencing Kit V4
is a gel-free solution that was shown previously to produce high rates of miRNA mapping and discovery when working with low input samples such as plasma and serum. Here we wanted to verify that NEXTFLEX® Small RNA-Seq Kit can also be used to investigate microRNAs in a population of purified exosomes. Additionally, the protocol is completely automated protocol from library prep to normalization pooling on the Sciclone® G3 NGS/NGSx workstations
NEXTFLEX Small RNA Sequencing Kit V4
is a gel-free solution that was shown previously to produce high rates of miRNA mapping and discovery when working with low input samples such as plasma and serum. Here we wanted to verify that NEXTFLEX® Small RNA-Seq Kit can also be used to investigate microRNAs in a population of purified exosomes. Additionally, the protocol is completely automated protocol from library prep to normalization pooling on the Sciclone® G3 NGS/NGSx workstations
 Sciclone G3 NGSx iQ Workstation
and Zephyr® G3 NGS workstations
Sciclone G3 NGSx iQ Workstation
and Zephyr® G3 NGS workstations
 Zephyr G3 NGS Workstation
.
Zephyr G3 NGS Workstation
.

Methods
Exosomes from pooled human serum healthy donors and human bone marrow were obtained from System Biosciences. From each sample, 10 µg of purified exosomes was extracted using the NextPrep™ Magnazol™ cfRNA Isolation Kit from Revvity. This corresponds to approximately 2x 109 vesicles5.
12 µL of purified RNA were split into triplicate for sample preparation, equivalent to roughly to 10 pg of input of microRNA per library if we assume 1 miRNA/100 exosomes.
Small RNA libraries were prepared manually using the NEXTFLEX® Small RNA-Seq Kit v4 according to the manufacturer’s instructions except the adapters were used at ¼ dilution. Once libraries were prepared, they were quantified with Thermo Fisher® Scientific Qubit® fluorometer, pooled, and run on an Illumina® MiSeq® platform at 1x75 bp read lengths. Small RNA analysis was performed using a Revvity custom script. Alignment reference was mature miRNA from mirBase v22.1.
Results
The NEXTFLEX® Small RNA-Seq Kit v4 was able to generate gel-free libraries for all samples. After sequencing, filtering, and mapping the data, the proportion of reads that aligned with adapter dimer, tRNA, YRNA, rRNA and miRNA for both sample types were determined. Reads mapping to adapter dimer were below 3% in all cases

Figure 1: Average number of unique microRNA discovered from each sample
Unique miRNA species were identified and quantified in each sample at different thresholds (Figure 1). Even with the relatively low sequencing depth used in this experiment the diversity of the miRNA found in the human serum samples are in agreement with the values reported in the literature6.

Figure 2: Average top 10 miRNA observed with exosomes from bone marrow and pooled serum samples.
Finally, we looked at the top 10 miRNA expressed on each of the replicates of the exosome samples, to illustrate the differences in the content of each type of exosome (Figure 2).
Conclusion
In the present study, we presented a simplified, commercially available protocol encompassing exoRNA extraction and preparation of exosomal miRNA-Seq libraries from serum and bone marrow. This convenient gel-free workflow enables the construction of libraries from exoRNA. Using this workflow, researchers can achieve a high number of reads aligning to mature miRNA and low adapter dimers, even with the very low miRNA inputs characteristic of exosome samples.
References
- Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int J Nanomedicine. 2020 Sep 22;15:6917-6934.
doi: 10.2147/IJN.S264498. PMID: 33061359;
PMCID: PMC7519827. - Yang Y, Huang H, Li Y. Roles of exosomes and exosomederived miRNAs in pulmonary fibrosis. Front Pharmacol. 2022 Aug 11;13:928933. doi: 10.3389/fphar.2022.928933.
PMID: 36034858; PMCID: PMC9403513. - Chuanyun Li, Tong Zhou, Jing Chen, Rong Li, Huan Chen, Shumin Luo, Dexi Chen, Cao Cai, Weihua Li, The role of Exosomal miRNAs in cancer, Journal of Translational Medicine, 20, 1, (2022). https://doi.org/10.1186/s12967- 021-03215-4.
- Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14888-93. doi: 10.1073/pnas.1408301111.
Epub 2014 Sep 29. PMID: 25267620;
PMCID: PMC4205618. - xosome Research | System Biosciences [www.systembio.com/products/exosome-research]
- Shi H, Jiang X, Xu C, Cheng Q. MicroRNAs in Serum Exosomes as Circulating Biomarkers for Postmenopausal Osteoporosis. Front Endocrinol (Lausanne). 2022 Mar 10;13:819056. doi: 10.3389/fendo.2022.819056.
PMID: 35360081; PMCID: PMC8960856.
Featured products

NEXTFLEX Small RNA Sequencing Kit V4
The NEXTFLEX small RNA sequencing kit v4 uses patent-pending technology to provide a completely gel-free small RNA library preparation solution for Illumina and Element sequencing platforms.
- Completely gel-free protocol from purified miRNA or as little as 1 ng total RNA, no PAGE cleanup required
- Automation-ready on Sciclone™ G3 NGSx, Sciclone G3 NGSx iQ™, and Zephyr™ G3 NGS workstations for walk-away scalability
- Low-bias ligation plus dimer reduction chemistry improve data quality from challenging biofluids (plasma, serum, urine, CSF) and extracellular vesicles.
- Internal benchmarking shows up to a 45% increase in unique miRNA species detected from as little as 1 ng of total RNA, supporting robust differential-expression analysis and novel small RNA discovery
- Libraries ready in ~ 6 hours, supporting rapid biomarker and functional genomics studies
- Includes 384 Unique Dual Index (UDI) barcodes for high-throughput multiplexing and minimal index hopping on Illumina®/Element® platforms
- Quality assured through rigorous testing of every lot
Explore our Dharmacon™ miRNA modulation reagents to enhance or inhibit miRNA levels as part of your functional studies.

Sciclone G3 NGSx Workstation with UV Light
The Sciclone™ G3 workstation can be configured to automate many automated liquid handling applications. Its open-deck design can incorporate a 96-or a 384-channel pipetting head, an optional eight-channel pipettor, and/or a bulk reagent dispenser. Revvity's "PING!" non-contact liquid level detection technology ensures that all samples are processed using vacuum or pressure filtration. Magnetic bead-based separations and a large variety of on-deck shaking, heating and cooling options are also supported with this liquid handling instrument.
Features/Benefits
- Open, modular design pipetting heads
- 96-and 384-well plate densities
- Volumes from 100 nL to 200 μL
- Independent 8-channel pipetting capability
- Compatible with a wide variety of options including a Sciclone gripper, bulk reagent modules, an on-deck microplate shaker, temperature-controlled locators, and a positive-pressure filtration system
- Hundreds of different possible configurations
We recommend using Revvity branded pipette tips with your liquid handler. Our tips have been carefully designed and produced to stringent standards of quality to ensure maximum accuracy and precision, and are the only tips guaranteed to work on Revvity liquid handlers.
Add a Z-8 pipetting module to your Sciclone liquid handler to enable cherry-picking, reformatting, normalization protocols and more. It provides eight channels with independent Z-axis (up and down) movement control and can aspirate and dispense different volumes at the same time.

Zephyr G3 NGS Workstation
The Zephyr G3 NGS liquid handling platform is a compact, easy-to-use system specifically designed for complete library prep automation or for the separation of pre- and post-PCR processes. It can handle up to 48 to 96 samples per day.
This workstation includes:
- Precise 96-channel pipetting head
- Integrated offset gripper enables consumable deck re-location and the ability to hold lids while pipetting (minimizing possible contamination)
- Partial-tip loading capability preventing excess tip waste
- On-deck temperature control options
- On-deck shaking options
- Waste Tip chute
- Maestro™ control software with Application Assistant User Interface
This workstation can prepare DNA, RNA, exome or metagenomics NGS library preparations and comes with over 40 verified library preparation methods.
We recommend using Revvity branded pipette tips with your liquid handler. Our tips have been carefully designed and produced to stringent standards of quality to ensure maximum accuracy and precision, and are the only tips guaranteed to work on Revvity liquid handlers.
Featured categories

miRNA Analysis
The purification of small RNA species such as microRNA (miRNA) is challenging because of the low concentrations, small fragment sizes (19-22 nucleotides) and tendency to be found in protein complexes.
Exosome/cfRNA Analysis
Extracellular vesicles are membrane-bound compartments which are being increasingly recognized as important players in cell-to-cell communication.

miRNA-seq Analysis
There are different species of small non-coding RNAs (sncRNAs) that play critical roles in various regulatory processes such as transcription, post-transcription, and translation.




























