
HTRF Human and Mouse Total LRRK2 Detection Kit, 500 assay points





| Feature | Specification |
|---|---|
| Application | Cell Signaling |
| Sample Volume | 16 µL |









Product information
Overview
Leucine-Rich Repeat Kinase 2 (LRRK2) is a multi-domain protein that phosphorylates a subset of Rab GTPases. The LRRK2/Rab pathway operates at the interface of vesicular trafficking, lysosomal functioning, and autophagy, and thus plays a crucial role in immune response and neuroinflammation. Ser1292 autophosphorylation serves as an indicator of LRRK2 kinase activity. Pathogenic LRRK2 mutations, such as the common mutation G2019S, increase LRRK2 kinase activity, resulting in a toxic hyperactive protein that contributes to the Parkinson’s Disease (PD) phenotype. Consequently, LRRK2 inhibitors and degraders are considered promising therapeutic agents.
How it works
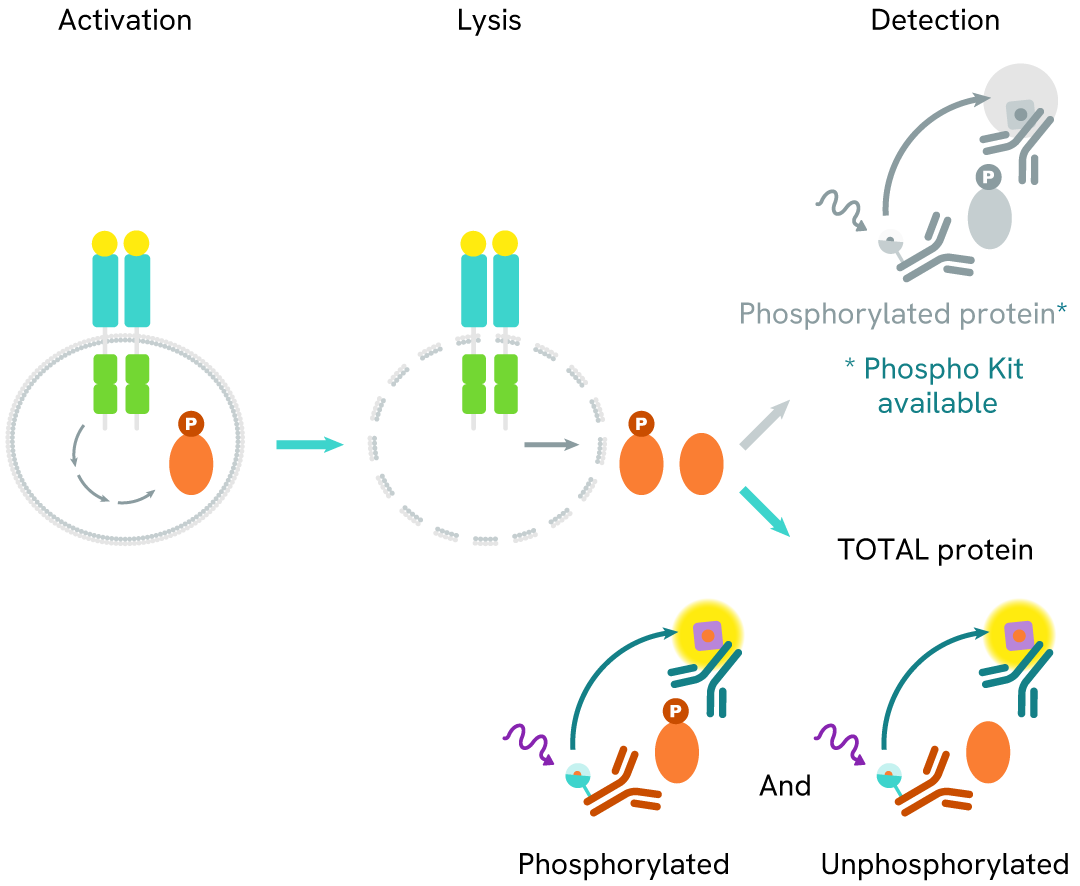
Total LRRK2 assay principle
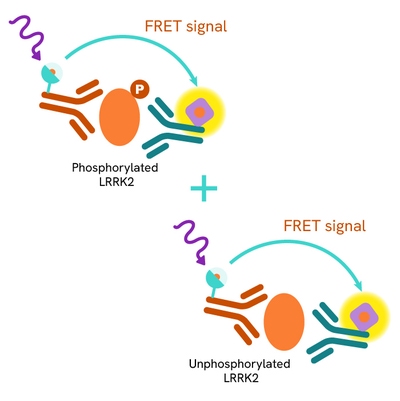
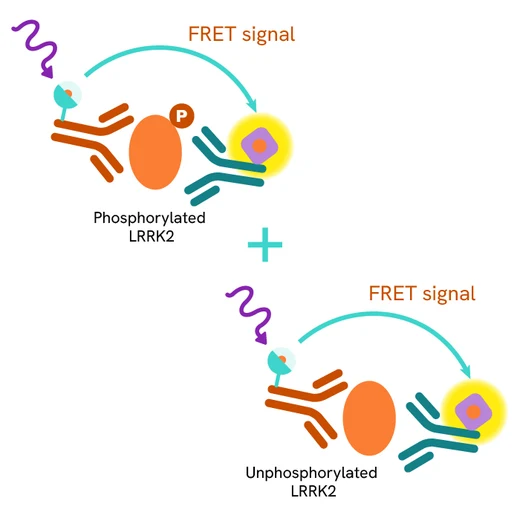
The Total LRRK2 assay quantifies the expression level of LRRK2 in a cell lysate. Unlike Western Blot, the assay is entirely plate-based and does not require gels, electrophoresis, or transfer. The Total LRRK2 assay uses two labeled antibodies, one coupled to a donor fluorophore and the other to an acceptor. Both antibodies are highly specific for a distinct epitope on the protein. In the presence of LRRK2 in a cell extract, the addition of these conjugates brings the donor fluorophore into close proximity with the acceptor and thereby generates a FRET signal. Its intensity is directly proportional to the concentration of the protein present in the sample and provides a means of assessing the protein's expression under a no-wash assay format.

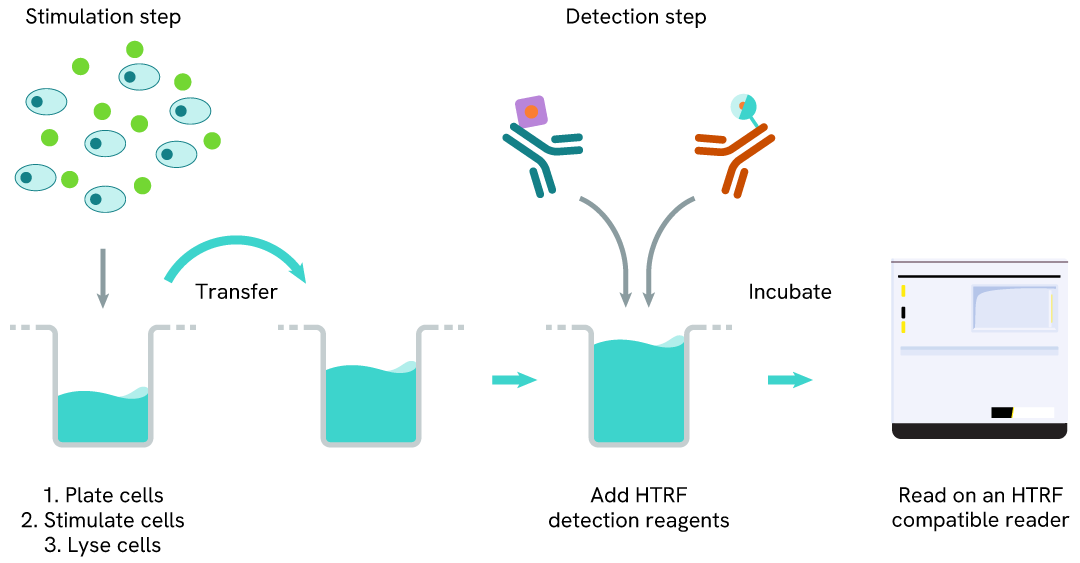
Total LRRK2 two-plate assay protocol
The two-plate protocol involves culturing cells in a 96-well plate before lysis, then transferring lysates into a 384-well low volume detection plate before the addition of HTRF Total LRRK2 detection reagents. This protocol enables the cells' viability and confluence to be monitored.
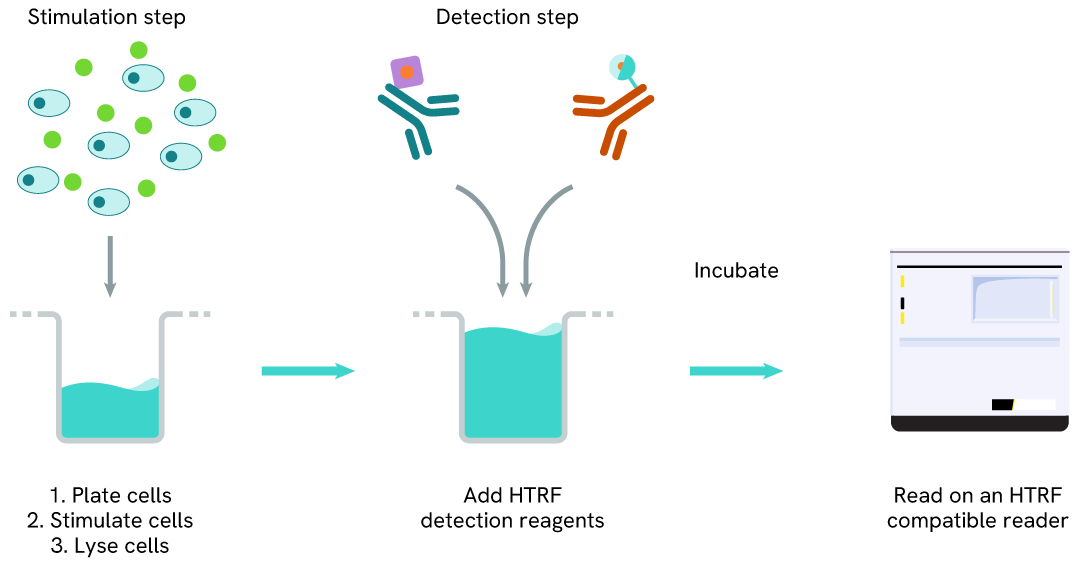
Total LRRK2 one-plate assay protocol
Detection of Total LRRK2 with HTRF reagents can be performed in a single plate used for culturing, stimulation, and lysis. No washing steps are required. This HTS designed protocol allows miniaturization while maintaining robust HTRF quality.
Assay validation
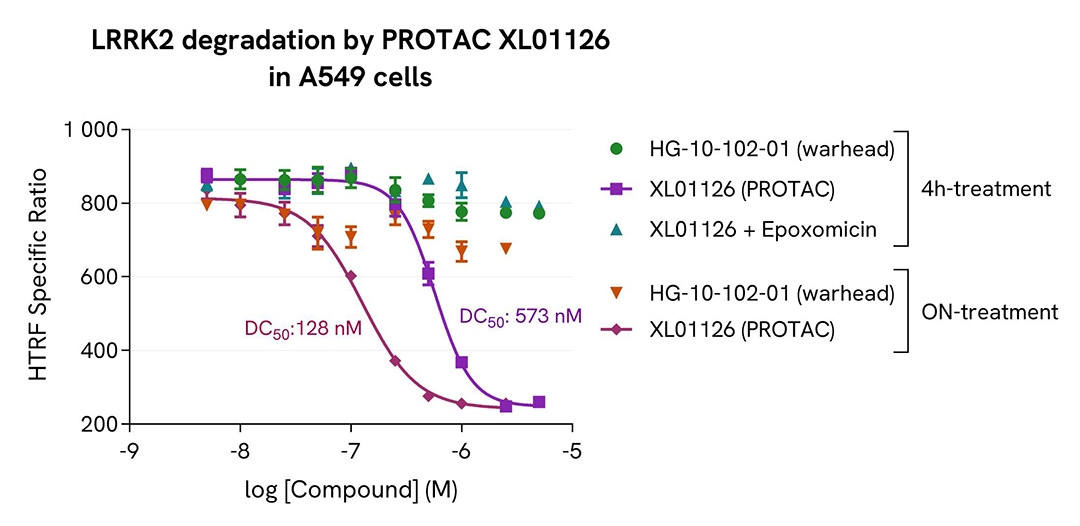
Degradation of LRRK2 by PROTAC XL01126
The experiments were carried out according to the two-plate assay protocol on adherent cells. A549 cells were plated in a 96-well culture plate (100,000 cells/well) and incubated for 5 hours at 37°C, 5% CO2. The cells were treated with increasing doses of LRRK2 PROTAC XL01126 or with the warhead HG-10-102-01 (negative control) for 4 hours or overnight. The 4-hour treatment with the PROTAC was performed in the absence or presence of 1 µM Epoxomicin (proteasome inhibitor). At the end of the treatment, the cells were lysed with 50 µL of supplemented lysis buffer #4, and 16 µL of lysate were transferred into a 384-well low volume white microplate (ProxiPlate-384 Plus, Cat # 6008280/9) before the addition of 4 µL of the HTRF Total LRRK2 detection antibodies. The HTRF signal was recorded on an EnVision Nexus reader after an incubation step of 2 hours at room temperature. Cell viability was also assessed by transferring 5 µL of the same lysate into an HTRF 96-well low volume white plate (Cat # 66PL96005/025/100), followed by the addition of 25 µL of ATPlite 1step reagent (ATPlite 1step Luminescence Assay System, Cat # 6016736/1/9). The luminescence signal was measured on an EnVision Nexus reader after a 10-minute incubation in the dark.
As expected, the warhead HG-10-102-01 did not induce any signal decrease, while PROTAC XL01126 dose-dependently degraded LRRK2. This degradation was blocked in the presence of Epoxomicin. In this experiment, the overnight treatment with the PROTAC yielded the best DC50 (4.5 times better than that obtained after a 4-hour treatment). The highest degradation rate (Dmax) was approximately 70% and was achieved with 1 µM of PROTAC. These results were further supported by the ATPlite 1step viability assay, where no cytotoxic effects were observed for the different compounds (data not shown here)
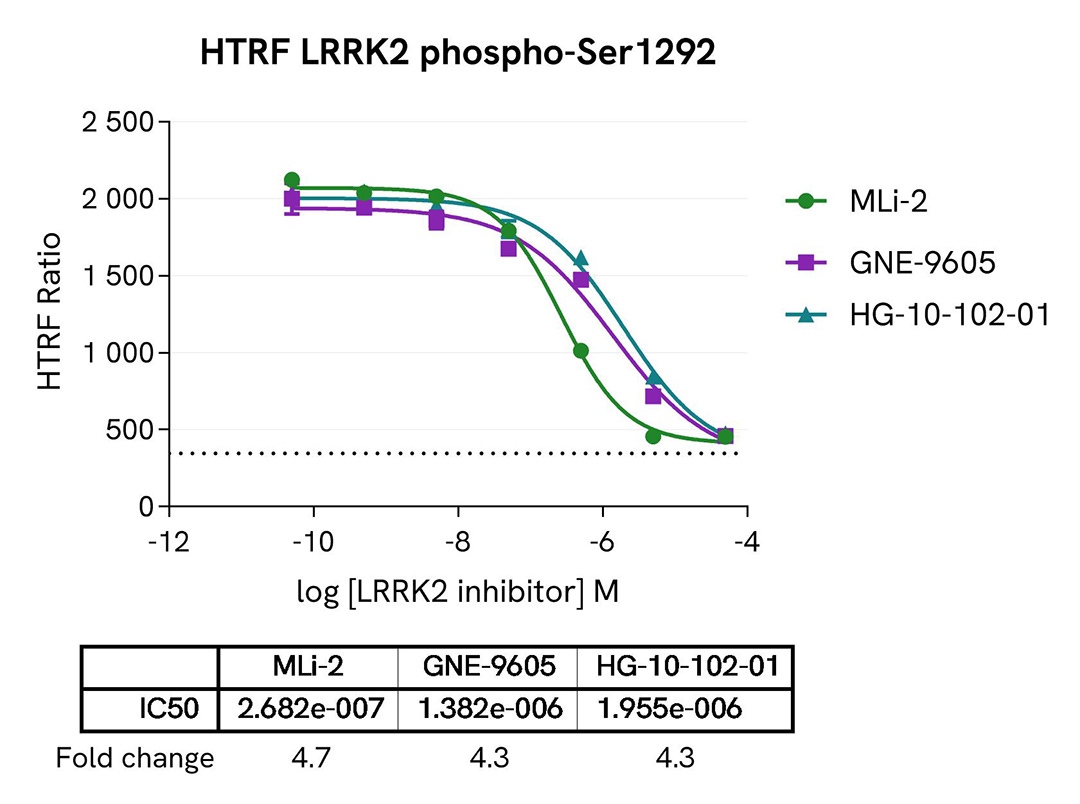
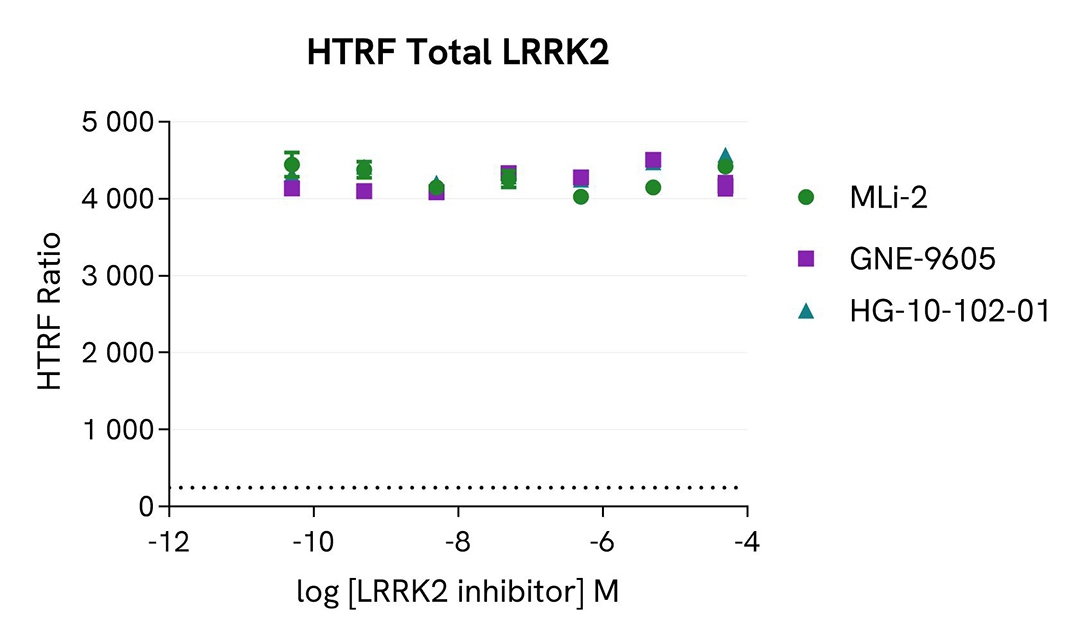
Inhibition of LRRK2 Ser1292 autophosphorylation by LRRK2 inhibitors
HEK293 cells were seeded in a 96-well culture plate (50,000 cells/well) and transfected for 24 hours with a plasmid encoding the human mutant LRRK2 G2019S protein. The cells were treated for 4 hours with increasing concentrations of the LRRK2 inhibitors MLi-2, GNE-9605, and HG-10-102-01, and then incubated for 30 minutes with 50 nM Calyculin-A. The cells were lysed with 50 µL of supplemented lysis buffer #4 for 30 minutes at room temperature under gentle shaking.
For the detection of phospho-LRRK2 (Ser1292), 14 µL of lysate were transferred into a 384-well low volume white microplate and supplemented with 2 µL Activation buffer before the addition of 4 µL of the HTRF Phospho-LRRK2 (Ser1292) detection reagents. For the detection of total LRRK2, 16 µL of lysate (previously diluted 50-fold in supplemented lysis buffer) were transferred into the same detection plate, and 4 µL of the HTRF Total LRRK2 detection reagents were added. The HTRF signal was recorded on an EnVision Nexus reader after the incubation time recommended for each assay (overnight for Phospho-LRRK2 and 2 hours for Total LRRK2). Cell viability was also assessed using the ATPlite 1step Luminescence Assay System (Cat # 6016736/1/9) as described in the previous section.
As expected, the three inhibitors induced a dose-dependent decrease in LRRK2 autophosphorylation at Ser1292, while the expression level of LRRK2 remained unchanged. The cell viability monitored with the ATPlite 1step luminescence assay kit (Cat # 6016736/1/9) was not affected by the treatment with the inhibitors (data not shown here).
Detection of Total LRRK2 in different cell models
HEK293 cells were seeded in a 96-well culture plate at 12,500 cells/well and transfected with a plasmid encoding the human mutant LRRK2 G2019S protein. The human lung cancer cell line A549 and the mouse fibroblastic cell line NIH-3T3 were seeded at 100,000 and 200,000 cells/well, respectively, in a 96-well culture plate. After 24 hours of incubation, the cells were lysed with 50 µL of supplemented lysis buffer #4 for 30 minutes at room temperature under gentle shaking.
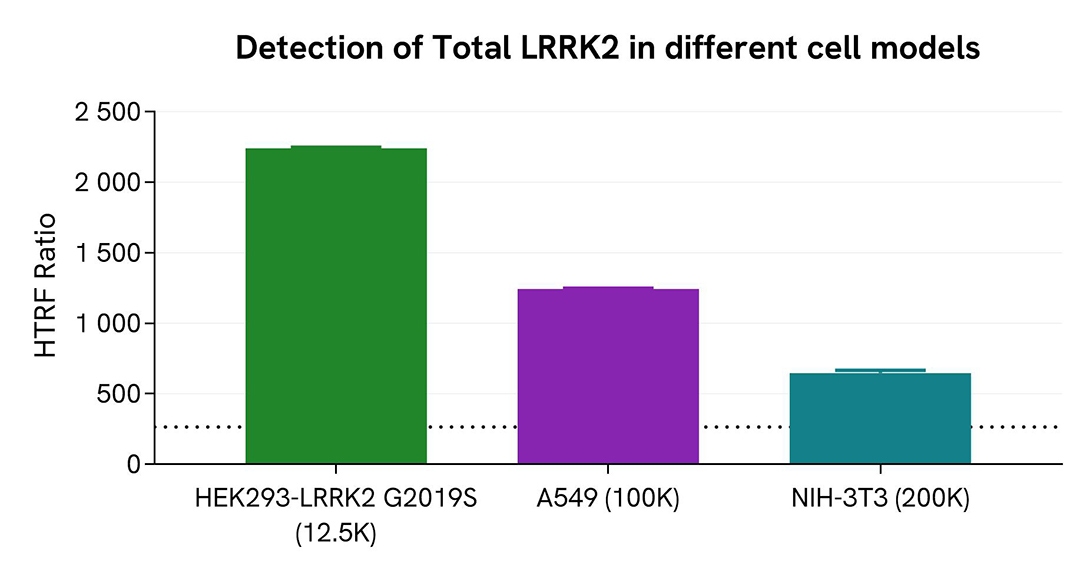
For the detection step, 16 µL of lysate were transferred into a 384-well low volume white microplate, and 4 µL of the HTRF Total LRRK2 detection reagents were added. The HTRF signal was recorded on an EnVision Nexus reader after 2 hours of incubation at room temperature.
The HTRF Total LRRK2 assay efficiently detected human LRRK2 G2019S in a cell model overexpressing the mutant protein and also enabled the detection of endogenous levels of the wild-type protein in human and mouse cell lines.
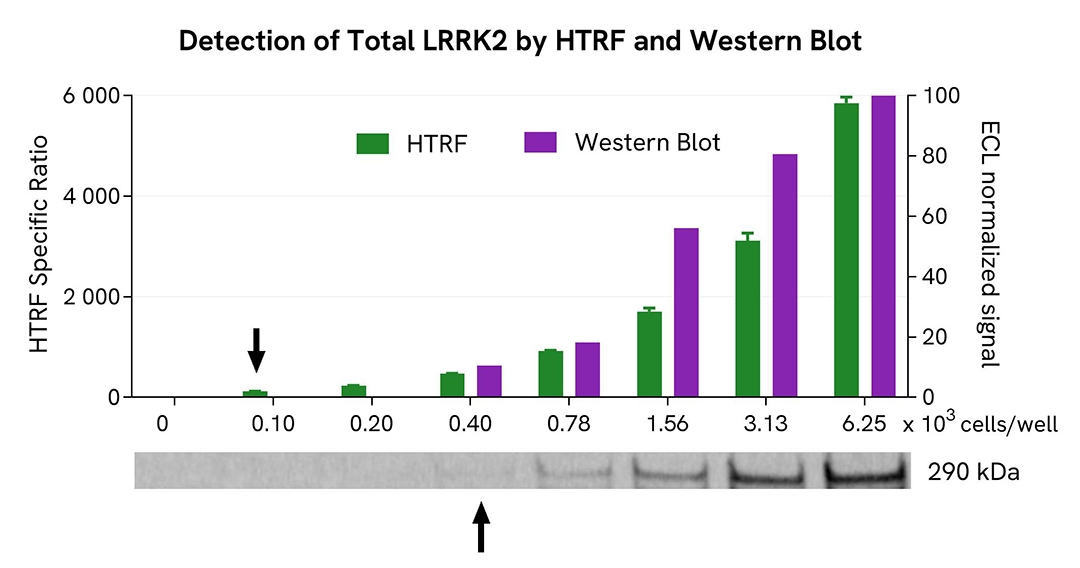
HTRF Total LRRK2 assay compared to Western Blot
HEK293 cells were transfected in a T175 flask with a plasmid encoding the human mutant LRRK2 G2019S protein for 24 hours, and then treated with 100 nM Calyculin-A for 30 minutes. The cells were lysed with 3 mL of supplemented lysis buffer #4 for 30 minutes at room temperature under gentle shaking.
Serial dilutions of the cell lysate were performed using supplemented lysis buffer, and 16 µL of each dilution were transferred into a 384-well low volume white microplate before the addition of 4 µL of HTRF Total LRRK2 detection reagents. Equal amounts of lysates were used for a side-by-side comparison between HTRF and Western Blot.
Using the HTRF Total LRRK2 assay, 100 cells/well were enough to detect a significant signal, while 400 cells were needed to obtain a minimal chemiluminescent signal using Western Blot. Therefore, under these conditions, the HTRF Total LRRK2 assay was four times more sensitive than the Western Blot technique.
Simplified pathway
LRRK2 signaling pathway
Under physiological conditions, LRRK2 activation and localization are regulated by the Rab GTPase Rab29. Rab29 recruits LRRK2 to the membranes of different organelles (trans-Golgi network, mitochondria, phagosomes, and damaged lysosomes) under specific stimuli. This recruitment leads to the stimulation of LRRK2 kinase activity and the subsequent increase in LRRK2 autophosphorylation at Ser1292. Activated LRRK2 phosphorylates a subset of membrane-associated Rab GTPases (Rab3, Rab8, Rab10, Rab12, Rab35, Rab43) and interacts with other downstream proteins, which together regulate a variety of cellular processes such as endocytosis and vesicular trafficking, microtubule dynamics, the autophagy-lysosomal pathway (ALP), Golgi and mitochondrial function, as well as α-synuclein homeostasis.
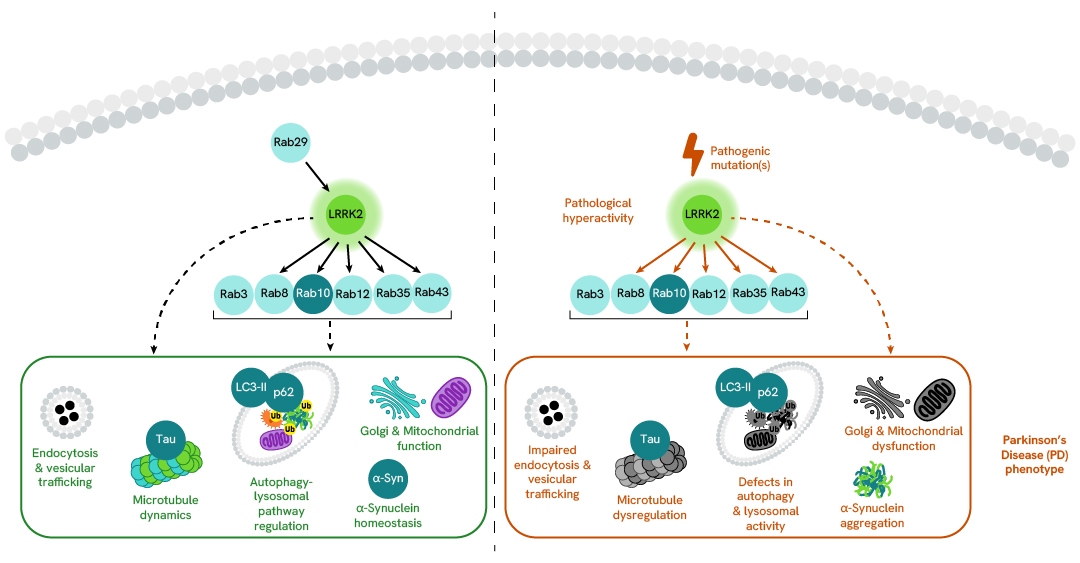
Pathogenic LRRK2 mutations, such as the common G2019S mutation located in the kinase domain, increase LRRK2 kinase activity, resulting in a toxic hyperactive protein that contributes to the Parkinson’s Disease (PD) phenotype.
Specifications
| Application |
Cell Signaling
|
|---|---|
| Automation Compatible |
Yes
|
| Brand |
HTRF
|
| Detection Modality |
HTRF
|
| Lysis Buffer Compatibility |
Lysis Buffer 4
|
| Molecular Modification |
Total
|
| Product Group |
Kit
|
| Sample Volume |
16 µL
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Target |
LRRK2
|
| Target Class |
Phosphoproteins
|
| Target Species |
Human
Mouse
|
| Technology |
TR-FRET
|
| Therapeutic Area |
Neuroscience
|
| Unit Size |
500 assay points
|
Video gallery
Resources
Are you looking for resources, click on the resource type to explore further.
Unlock the potential of HTRF™ LRRK2 immunoassays to support and advance Parkinson's disease drug discovery. This comprehensive...
Targeted protein degradation (TPD) offers a promising approach for treating neurodegenerative diseases by selectively degrading...


Loading...
How can we help you?
We are here to answer your questions.






























