

HTRF Human and Mouse Total ATG16L1 Detection Kit, 500 Assay Points





| Feature | Specification |
|---|---|
| Application | Cell Signaling |
| Sample Volume | 16 µL |









Product information
Overview
Autophagy Related 16 Like 1 (ATG16L1) is a protein that is essential for autophagy. ATG16L1 is recruited to the phagophore and recruits ATG5-ATG12 complex to a phagophore, facilitating LC3 lipidation (LC3-I to LC3-II conversion) and contributing to autophagosome maturation.
Phosphorylated ATG16L1 at Ser278 is only present on newly-forming autophagosomes, making its quantification an ideal surrogate for autophagic vesicle formation and autophagy induction. ATG16L1 is involved in conditions such as Alzheimer's Disease, colorectal cancer, heart disease, breast cancer, and chronic myeloid leukemia.
How it works
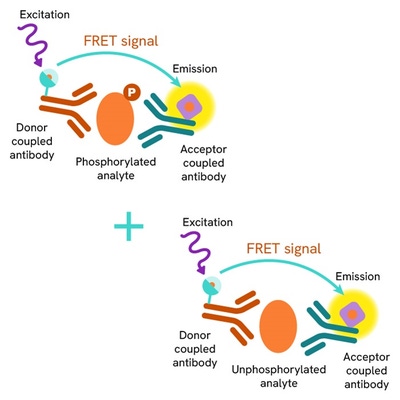
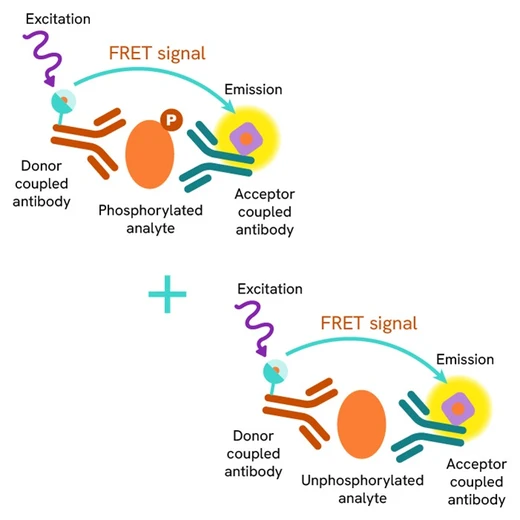
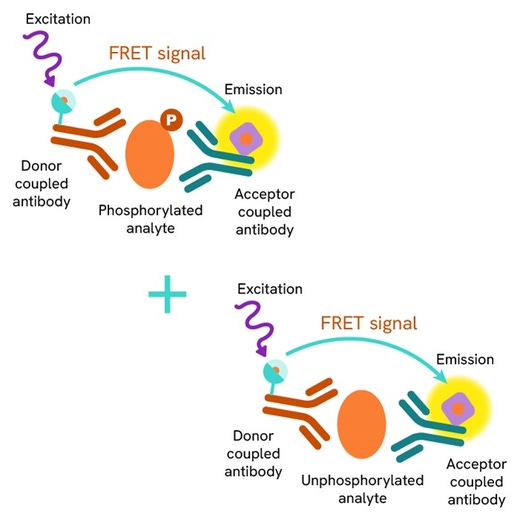
HTRF Total ATG16L1 assay principle
The HTRF Total ATG16L1 assay quantifies the expression level of ATG16L1 in a cell lysate. Unlike Western Blot, the assay is entirely plate-based, and does not require gels, electrophoresis, or transfer. The HTRF Total ATG16L1 assay uses two labeled antibodies, one coupled to a donor fluorophore, the other to an acceptor. Both antibodies are highly specific for a distinct epitope on the protein. In presence of ATG16L1 in a cell extract, the addition of these conjugates brings the donor fluorophore into close proximity with the acceptor, and thereby generates a FRET signal. Its intensity is directly proportional to the concentration of the protein present in the sample, and provides a means of assessing the protein’s expression under a no-wash assay format.

HTRF Total ATG16L1 two-plate assay protocol
The two-plate protocol involves culturing cells in a 96-well plate before lysis, then transferring lysates into a low volume detection plate (either HTRF 384-lv or 96-lv plate) before the addition of HTRF Total ATG16L1 detection reagents. This protocol enables the cells' viability and confluence to be monitored

HTRF Total ATG16L1 one-plate assay protocol
Detection of Total ATG16L1 with HTRF reagents can be performed in a single plate used for culturing, stimulation, and lysis. No washing steps are required. This HTS designed protocol enables miniaturization, while maintaining robust HTRF quality.

Assay validation
Validation of HTRF total ATG16L1 and Phospho-ATG16L1 (Ser278) kits on the human U-87 MG cell line using mTOR and ULK1/2 inhibitors
Human U-87 MG cells were plated in a 96-well plate (100,000 cells/well) in complete culture medium, and incubated overnight at 37°C, 5% CO2. The cells were treated with a dose-response of AZD2014, an mTOR inhibitor, for 3 h at 37°C, 5% CO2. Following culture medium removal, cells were lysed with 25 µl of supplemented lysis buffer #3 (1X) + BR (2X) for 1 h at RT under gentle shaking. After cell lysis, 16 µL of lysate were transferred into a 384-well low volume white microplate, then 4 µL of the HTRF Phospho-ATG16L1 (Ser 278) or Total ATG16L1 detection reagents were added. The HTRF signals for the two kits were recorded after an overnight incubation at room temperature. As expected, AZD2014, a potent mTOR inhibitor, induced autophagy activation, leading to a dose-dependent increase in ATG16L1 phosphorylation on Serine 278 without any significant effect on the expression level of the total ATG16L1 protein. Validation of HTRF Total ATG16L1 and Phospho-ATG16L1 (Ser278) kits on the human U-87 MG cell line using mTOR and ULK1/2 inhibitors Human U-87 MG cells were plated in a 96-well plate (100,000 cells/well) in complete culture medium, and incubated overnight at 37°C, 5% CO2. The cells were treated with 2.5 µM of AZD2014, an mTOR inhibitor, and a dose-response of ULK-101, an autophagy kinase ULK1/2 inhibitor, for 3 h at 37°C, 5% CO2. Following culture medium removal, cells were lysed with 25 µl of supplemented lysis buffer #3 (1X) + BR (2X) for 1 h at RT under gentle shaking. After cell lysis, 16 µL of lysate were transferred into a 384-well low volume white microplate, then 4 µL of the HTRF Phospho-ATG16L1 (Ser 278) or Total ATG16L1 detection reagents were added. The HTRF signals for the two kits were recorded after an overnight incubation at room temperature. As expected, ULK-101, a potent and dual autophagy kinase ULK1/2 inhibitor, repressed autophagy activation induced by AZD2014, leading to a dose-dependent decrease in ATG16L1 phosphorylation on Serine 278, without any significant effect on the expression level of the ATG16L1 total protein.


Specificity of HTRF Total ATG16 assay using HAP1 ATG16 knock-out cells
The human total ATG16L1 expression level was assessed using the HTRF Total kit in Wild Type (WT) and ATG16 knock-out (KO) HAP1 cells. The cell lines were cultured in flasks and incubated 48 h at 37°C, 5% CO2. After culture medium removal, cells were lysed with supplemented lysis buffer #3 (1X) + BR (2X) for 30 min at RT under gentle shaking. After cell lysis, 16 µL of lysate were transferred into a 384-well low volume white microplate then 4 µL of the HTRF Total ATG16L1 detection reagents were added. In parallel, GAPDH level was monitor using the GAPDH Housekeeping Cellular Kit (# 64GAPDHPET/G/H). 4 µL of cell lysate were prediluted with 180 µL of diluent #11. Then, 16 µL of this diluted material were mixed with 4 µL of the HTRF GAPDH Housekeeping Cellular detection reagents. HTRF signal for both kits was recorded after an overnight incubation at room temperature. The measured level of GAPDH was equivalent under both tested conditions. In contrast, while the HTRF Total ATG16L1 signal was significantly positive in HAP1 WT cells, it was dramatically decreased (equivalent to the negative signal) in HAP1 ATG16 KO cells, demonstrating the specificity of the HTRF Total ATG16L1 assay for the detection of the ATG16L1 protein. HAP1 ATG16 KO (10bp deletion) cell line from Horizon Discovery # HZGHC000296c010 HAP1 WT cell line from Horizon Discovery # C631

Versatility of HTRF Total ATG16L1 assay in cell lines from multiple species
Human (PC-3), mouse (C2C12) and rat (H4-II-E) cell lines were seeded at 100,000 cells/well in a 96-well plate. After a 24h incubation, the cells were lysed with 25 µL of supplemented lysis buffer #3 (1X) + BR (2X) for 1 h at RT under gentle shaking. After cell lysis, 16 µL of lysates were transferred into a 384-well low volume white microplate then 4 µL of the HTRF Total ATG16L1 detection reagents were added. The HTRF signal was recorded after an overnight incubation at room temperature. The HTRF Total ATG16L1 assay efficiently displayed significant positive signals in the cell lines from the 3 species tested, demonstrating its capability to detect human, mouse and rat versions of the protein, and its suitability for translational research.

HTRF Total ATG16L1 Detection Kit compared to Western Blot
HCT116 cells were cultured in flasks and incubated 48 h at 37°C, 5% CO2. Then, the cells were treated with 100 nM of Calyculin A for 10 min. After culture medium removal, cells were lysed with supplemented lysis buffer #3 (1X) + BR (2X) for 30 min at RT under gentle shaking. Soluble supernatants were collected after 10 min centrifuging. Equal amounts of lysates were used for a side-by-side comparison of WB and HTRF. The HTRF Total ATG16L1 assay is 4 times more sensitive than Western Blot assay.

Simplified pathway
Simplified ATG16 signaling pathway in macroautophagy
Autophagy is an evolutionary conserved cellular process that occurs in virtually all eukaryotic cells, ranging from yeast to mammals. Autophagy is crucial in the maintenance of homeostasis, being involved in numerous physiological processes including stress responses (e.g. starvation, hypoxia, high temperature), cell growth, and aging. Conversely, dysfunctions in autophagic mechanisms have been associated with diseases such as cancer, neurodegenerative diseases, infectious diseases, and cardiac and metabolic diseases. There are 3 types of autophagy: macroautophagy, microautophagy and chaperone-mediated autophagy. Macroautophagy (hereafter referred to as autophagy) starts with the formation of a bud from lipid phosphatidylinositol 3-phosphate rich membranes such as reticulum endoplasmic, Golgi, or plasma membranes, and known as a pre-autophagosomal structure (PAS). This cup-shaped structure requires the ULK1/Atg1 complex formed by Atg13, Atg101, FIP200, and ULK1 proteins. A transmembrane protein, Atg9, contained in vesicles is involved in the recruitment of Atg2-Atg18 to the PAS. With its phospholipid transfer activity, the Atg2 protein supplies phospholipids necessary for membrane elongation. ULK1 phosphorylates and activates Beclin1, which partners with Vsp15, Vsp34, Atg14, and NRFB2/Atg38 to form a class III Phosphatidylinositol 3-kinase (PI3KC3-Complex 1). This complex leads to the production of phospho-inositol triphosphate (PIP3), which further directs the recruitment of PI3P binding Atg18/WIPI 1–4 proteins and the Atg12-Atg5-Atg16 complex. The latter complex, along with Atg4 and Atg7, is implicated in the LC3B lipidation (LC3-I to LC3-II conversion) on the phagophore. This contributes to autophagosome maturation, which subsequently fuses with lysosomes forming an autolysosome, where trapped cellular components are degraded and building blocks recycled. Phosphorylated ATG16L1 at Ser278 is only present on newly-forming autophagosomes, making its quantification an ideal surrogate for autophagic vesicle formation and autophagy induction. Importantly, its levels are not affected by prolonged stress or late-stage autophagy blocks, which can jeopardize autophagy analysis. Moreover, ATG16L1 phosphorylation by the autophagy kinase ULK1 is a conserved indicator of autophagy induction, which is activated by multiple stimuli. Taken together, analysis of ATG16L1 phosphorylation represents an excellent readout when studying autophagy induction.

Specifications
| Application |
Cell Signaling
|
|---|---|
| Brand |
HTRF
|
| Detection Modality |
HTRF
|
| Lysis Buffer Compatibility |
Lysis Buffer 1
Lysis Buffer 3
Lysis Buffer 4
|
| Molecular Modification |
Total
|
| Product Group |
Kit
|
| Sample Volume |
16 µL
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Target Class |
Phosphoproteins
|
| Target Species |
Human
Mouse
|
| Technology |
TR-FRET
|
| Unit Size |
500 assay points
|
Video gallery
Resources
Are you looking for resources, click on the resource type to explore further.
Discover the versatility and precision of Homogeneous Time-Resolved Fluorescence (HTRF) technology. Our HTRF portfolio offers a...
This guide provides you an overview of HTRF applications in several therapeutic areas.
Neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, and Huntington’s, are complex disorders that affect millions...


Loading...
How can we help you?
We are here to answer your questions.






























