

HTRF EPIgeneous Binding Domain A Detection Kit, 500 Assay Points





| Feature | Specification |
|---|---|
| Application | Biochemical Enzymatic Assay |
| Sample Volume | 120 µL |









Product information
Overview
The EPIgeneous Binding Domain kit series provides a simple biochemical approach to study epigenetic reader domain interactions with modified histones. All kits are based on a GST-tagged binding domain / biotin-coupled Histone peptide format, and can be run using the same add-and-read single plate protocol.
Binding Domain kit A has been successfully validated on 4 different domains, including some important therapeutic targets, in the BRD family.
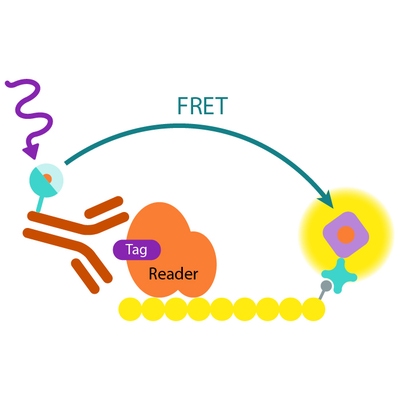
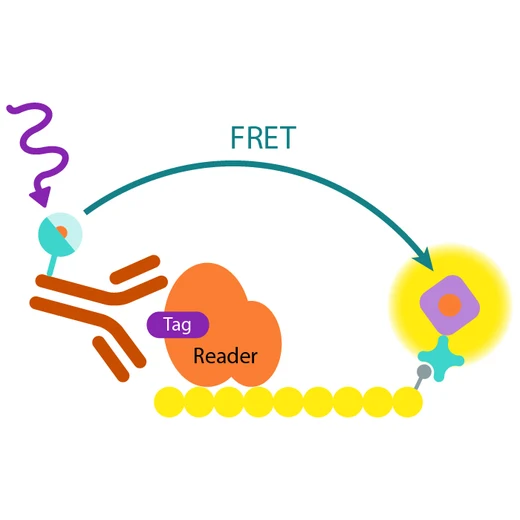
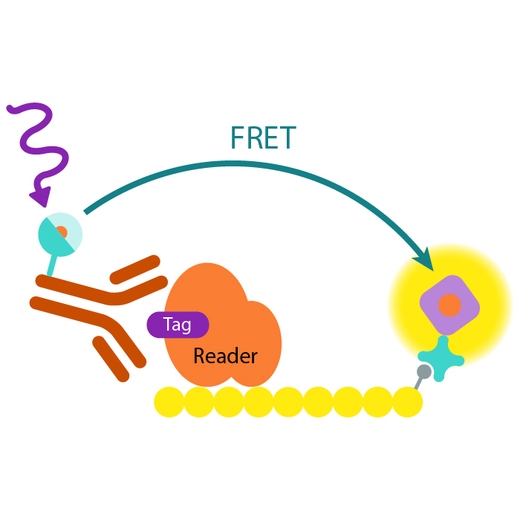
How it works
Assay principle
In EPIgeneous Binding Domain kit A, the GST-tagged reader domain protein binding to the biotinylated peptide substrate is detected by a conjugate mix: anti-GST Eu cryptate-labeled antibody conjugate (donor), and d2-conjugated streptavidin (acceptor). The interaction of the reader domain with the substrate brings the donor and acceptor dyes into close proximity, and allows FRET to occur upon light excitation. The specific signal at 665 nm is inhibited when a specific compound prevents the reader domain protein from binding to its substrate.

Analytical performance
Full assay optimization performed on BRD4(1) using EPIgeneous Binding Domain Kit A. Additional data describes the use of EPIgeneous Binding Domain Kit A for the profiling of reference (+)-JQ1 compound on multiple binding domains.
Peptide-biotin titration - BRD4(1) / histone H4 peptide interaction
The GST-Reader concentration was fixed at 5nMf while the peptide-biotin was serially diluted (100nMf to 0.005nMf).
For each peptide-biotin concentration, a negative control was performed by removing the GST-Reader protein from the wells. This negative control was used as non specific signal to calculate the HTRF delta ratio. This specific signal was proportional to the specific interaction measured between GST-BRD4(1) and [Lys(5,8,12,16)Ac]-H4(1-21)-biotin peptide.
The 4nM Kd value was determined from this experiment using a one-site specific binding regression (saturation equations).

DMSO tolerance - peptide-biotin concentration optimization
Peptide-biotin titration performed with various DMSO percentages. The apparent Kd values are determined using one-site specific binding regression (saturation equations). Due to the competitive inhibitor nature of the DMSO on the BRD4(1)/H4 peptide interaction (1), a shift of apparent Kd, is observed while DMSO% increases. As the DMSO competes on BRD4(1)/H4 peptide interaction, the assay window decreases while the DMSO percentage increases. The assay is then recovered by increasing the peptide-biotin concentration. In the case shown here, the optimal peptide-biotin concentration was set between the real Kd and the EC100 obtained for the titration without DMSO, a compromise between assay window and assay sensitivity to enable inhibitor studies (1% DMSO and 3nM peptide-biotin).


Inhibitor titration - BRD4(1) inhibition by reference compounds
The assay was performed using 3 nM peptide-biotin, 5 nM GST-BRD4(1) and 1% DMSO, all set constant throughout the inhibitor titration.
The IC50 of (+)-JQ1 and H4 tetra-acetylated peptide are in good agreement with published data (1, 2).

Z' Factor - Assay robustness
The assay was performed using 3 nM peptide-biotin, 5 nM GST-BRD4(1) and 1% DMSO.
The 0.76 Z' factor underlines the robustness of the assay and its suitability for HTS.

Compound profiling - Assay selectivity using various reader domain
(+)-JQ1 compound was profiled on the BET bromodomain family, CREBBP and BAZ2B bromodomains using EPIgeneous Binding Domain Kit A and B. As already described (3), (+)-JQ1 is a non-selective inhibitor over the BET family but displays selectivity over non BET bromodomains (CREBBP and BAZ2B).

Assay validation
Validated binding domains
Three kits (A, B and C) have already been validated and fully optimized on a selection of 28 key binding domains. For non-validated reader domains, a fourth one, the Discovery Kit, enables researchers to profile which of the A, B or C kits is the best assay solution.
| BINDING DOMAIN KIT A | BINDING DOMAIN KIT B | BINDING DOMAIN KIT C |
|---|---|---|
| BRD2(1) | CECR2 | BRD4(1/2) |
| BRD3(1) | FALZ (BPTF) | ATAD2A |
| BRD4(1) | BRD2(2) | ATAD2B |
| CBX 1 | BRD2(1/2) | BRD9 |
| BRD3(2) | SMARCA4 (BRG1) | |
| BRD3(1/2) | BAZ2B | |
| BRD4(2) | ||
| BRDT(1) | ||
| BRDT(1/2) | ||
| CREBBP | ||
| BRD1 | ||
| BRPF3 | ||
| TAF1L(2) | ||
| TAF1L(1/2) | ||
| TAF1(2) | ||
| TAF1(1/2) | ||
| L3MBTL1 | ||
| UHRF1 |
Specifications
| Application |
Biochemical Enzymatic Assay
|
|---|---|
| Brand |
EPIgeneous
|
| Detection Modality |
HTRF
|
| Product Group |
Kit
|
| Sample Volume |
120 µL
|
| Shipping Conditions |
Shipped in Dry Ice
|
| Target Class |
Epigenetics
|
| Technology |
TR-FRET
|
| Therapeutic Area |
Metabolism/Diabetes
Neuroscience
Oncology & Inflammation
|
| Unit Size |
500 assay points
|
Video gallery
Resources
Are you looking for resources, click on the resource type to explore further.
Discover the versatility and precision of Homogeneous Time-Resolved Fluorescence (HTRF) technology. Our HTRF portfolio offers a...
This guide provides you an overview of HTRF applications in several therapeutic areas.


Loading...
How can we help you?
We are here to answer your questions.






























