
NEXTFLEX Poly(A) Beads 2.0



The NEXTFLEX™ Poly(A) Beads 2.0 kit provides rapid, reliable mRNA enrichment by capturing polyadenylated RNA using oligo(dT) magnetic beads. This poly(A) selection step cleans up total RNA for high-quality RNA-seq library preparation, reducing rRNA carryover and background to improve transcript detection. The streamlined magnetic separation workflow supports consistent mRNA isolation across studies and integrates seamlessly upstream of all RNA sequencing library prep workflows including NEXTFLEX RNA-seq kits. Ideal for transcriptome profiling and differential expression studies where robust mRNA capture and reproducibility are critical.
| Feature | Specification |
|---|---|
| Automation Compatible | Yes |
| Product Group | mRNA Enrichment |
The NEXTFLEX™ Poly(A) Beads 2.0 kit provides rapid, reliable mRNA enrichment by capturing polyadenylated RNA using oligo(dT) magnetic beads. This poly(A) selection step cleans up total RNA for high-quality RNA-seq library preparation, reducing rRNA carryover and background to improve transcript detection. The streamlined magnetic separation workflow supports consistent mRNA isolation across studies and integrates seamlessly upstream of all RNA sequencing library prep workflows including NEXTFLEX RNA-seq kits. Ideal for transcriptome profiling and differential expression studies where robust mRNA capture and reproducibility are critical.



Product information
Overview
- High-efficiency mRNA enrichment – selectively captures polyadenylated mRNA to reduce rRNA/background and improve transcript detection for RNA-seq.
- Consistent, automation-friendly workflow – rapid magnetic bead protocol supports reproducible mRNA isolation across samples and batches.
- Seamless compatibility – Works with any RNA-seq library prep, regardless of sequencing platform downstream. Optimized for NEXTFLEX RNA-seq kits.
- Versatile input – from 10 ng to 5 µg of total RNA from any eukaryotic species can be used as starting material for transcriptome profiling and differential expression studies.
- Low hands-on time – simple magnetic separation helps reduce variability.
- Small-volume elution; no precipitation step, oligo(dT) magnetic bead capture elutes intact mRNA in low volumes, eliminating ethanol precipitation to save time and preserve yield, especially for low-input RNA.
Additional product information
Why poly(A) selection with oligo(dT) magnetic beads
For intact total RNA, poly(A) selection with oligo(dT) magnetic beads delivers focused mRNA enrichment that standardizes input for RNA-seq. This approach supports short-read and long-read bulk RNA-seq, 3′ mRNA-seq, transcriptome profiling, and differential expression workflows with a fast, automation-ready magnetic protocol. Use poly(A) selection when your study targets polyadenylated mRNA and RNA quality is high; choose rRNA depletion instead for degraded RNA or when interrogating non-polyadenylated transcripts (e.g., lncRNA), ensuring the right upstream cleanup for your downstream library prep.
For background on why enriching poly(A)+ transcripts improves RNA-seq signal, see our blog RNA-seq: a multilayered view of the mRNA lifecycle.

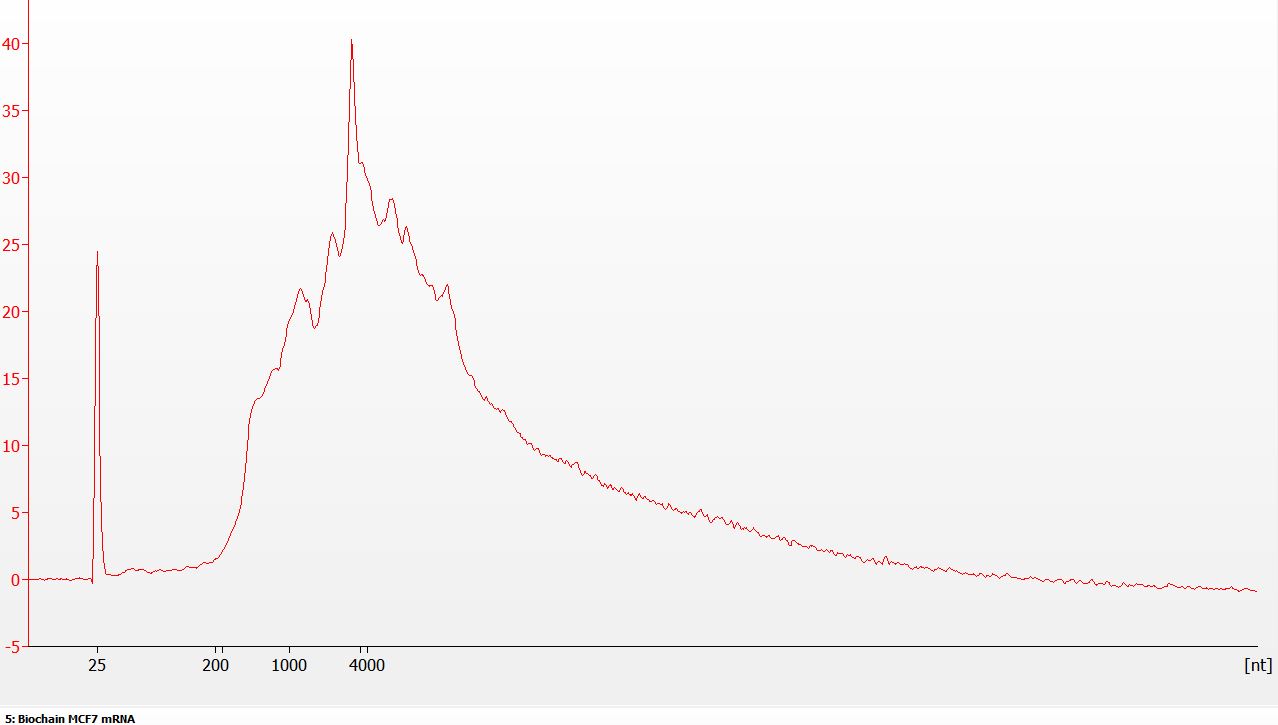
Figure 1: Representative Poly(A) Enrichment Profile. NEXTFLEX Poly(A) Beads 2.0 produce clean poly(A)+ mRNA with low residual rRNA. From 5 µg total RNA MCF-7 cells, ≈ 1.5% of RNA-seq reads mapped to rRNA, indicating effective removal of rRNA background prior to RNA-seq library preparation.
Reducing rRNA reads in RNA-seq libraries
Total RNA typically contains ~ 80–90% rRNA, so sequencing it directly wastes reads and compute. NEXTFLEX Poly(A) Beads 2.0 hybridize to 3′ poly(A) tails, enabling rapid magnetic separation that enriches polyadenylated mRNA and lowers rRNA/background before RNA-seq library preparation. The on-magnet workflow elutes in small volumes with no precipitation step required to preserve yield, support low-input RNA, and standardize mRNA isolation across samples for all compatible RNA-seq kits. The result is cleaner poly(A)+ mRNA and improved transcript detection for transcriptome profiling and differential expression.
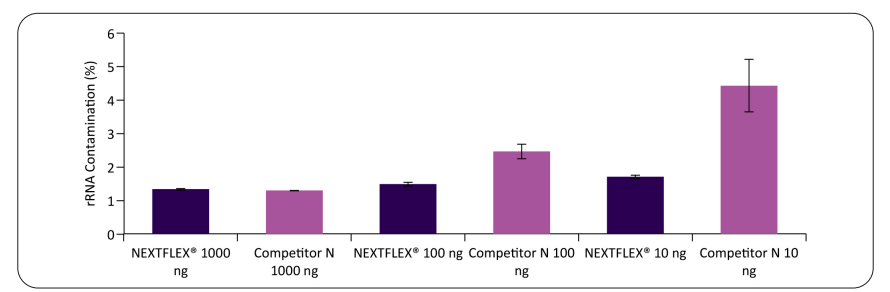
Figure 2: NEXTFLEX Poly(A) Beads 2.0 yield lower rRNA carryover across input amounts. Poly(A)+ mRNA was isolated from Universal Human Reference RNA (Agilent) using NEXTFLEX Poly(A) Beads 2.0 and a competitor poly(A) kit at 1000 ng, 100 ng, and 10 ng.
Seamless integration with NEXTFLEX RNA ecosystem
NEXTFLEX Poly(A) Beads 2.0 slot cleanly into Revvity’s broader NEXTFLEX RNA-seq accessory portfolio , letting you tailor each run to sample type and throughput. Other upstream modules include our ribodepletion portfolio . Downstream, you can scale from pilot sets to 384-plex studies with NEXTFLEX RNA-Seq 2.0 UDI Barcodes or use UDI-UMI adapter plates, adding molecular barcodes when duplicate suppression matters. Adapter lots are color-balanced and QC-verified for index purity, helping limit barcode bleed-through in large pools. Explore the full accessory line to build a single-vendor workflow that grows from low-input discovery to high-throughput studies.
Automation-scripts available
Pre-built scripts for Revvity workstations cut hands-on time e while preserving the metrics that matter the most. Ideal for labs balancing speed, reproducibility, and walk-away convenience.
Specifications
| Automation Compatible |
Yes
|
|---|---|
| Format |
Automation Friendly Volumes
|
| Product Group |
mRNA Enrichment
|
| Shipping Conditions |
Shipped Ambient
|
| Unit Size |
96 rxns
|
Citations
Plant transcriptomics
- Decsi, K., Ahmed, M., Rizk, R., Abdul-Hamid, D., Vaszily, Z., & Tóth, Z. (2024). Transcriptome datasets of maize plant cultures treated with humic- and amino acids. Data in Brief, 57, 110900. https://doi.org/10.1016/j.dib.2024.110900
Model organisms/fundamental biology
- Higdon, A. L., Won, N. H., & Brar, G. A. (2024). Truncated protein isoforms generate diversity of protein localization and function in yeast. Cell Systems, 15(4), 388–408.e4. https://doi.org/10.1016/j.cels.2024.03.005
FAQs
-
When should I choose poly(A) selection vs. rRNA depletion for RNA-seq?
-
What input amount does NEXTFLEX Poly(A) Beads 2.0 support?
-
Do I need an ethanol precipitation step after enrichment?
-
Is the kit compatible with automation platforms?
-
How does poly(A) selection reduce rRNA reads?
-
Has this kit been used with plant or non-mammalian RNA?
-
What magnetic stand do I need?
Resources
Are you looking for resources, click on the resource type to explore further.
This flyer describes the benefits of the NEXTFLEX® Rapid Directional RNA-seq 2.0 Kit.
This flyer illustrates the breadth of the NEXTFLEX RNA-seq Portfolio
Application note describing RNA-seq transcriptome profiling of five drug discovery cell lines to support model selection, pathway...


Loading...
How can we help you?
We are here to answer your questions.






























