
Simplified genome wide detection of genetic variants with NEXTFLEX Rapid XP V2 library prep kit
Whole Genome Sequencing (WGS) is a powerful tool that can detect a broad spectrum of genetic variations, from SNP to large structural variants, across the entire genome of an individual. With decreasing sequencing costs, WGS is becoming a viable option for population-wide screening for specific disease risk, including expanded carrier testing, pharmacogenomic variants, and polygenic risk scores, for example. Additionally, WGS is also gaining interest as a complementary research tool in the context of newborn screening, a cornerstone of pediatric healthcare.
To view the full content please answer a few questions
Introduction
Library prep can be intimidating for labs without extensive NGS experience. The NEXTFLEX™ Rapid XP V2
 NEXTFLEX Rapid XP V2 DNA-Seq Kit
Discover
Library prep kit incorporates a streamlined protocol to simplify library prep while delivering robust results. Pooling with the NEXTFLEX UDI barcodes
NEXTFLEX Rapid XP V2 DNA-Seq Kit
Discover
Library prep kit incorporates a streamlined protocol to simplify library prep while delivering robust results. Pooling with the NEXTFLEX UDI barcodes
 NEXTFLEX UDI Barcodes (10NT, 1-24)
Discover
enables extreme multiplexing to reduce sequencing costs. Additionally, a complete automated workflow is available for labs seeking to increase throughput and minimize human error. Using cell line-derived reference standards with known mutations, we conducted a proof-of-concept study to evaluate the suitability of the NEXTFLEX Rapid XP V2 library prep kit for detecting germline mutations. These standards offer a controlled means to assess the performance of our WGS workflow, confirming its ability to identify genetic variants in sample material. This study also highlights the potential of the NEXTFLEX Rapid XP V2 kit for comprehensive genomic analysis in research applications.
NEXTFLEX UDI Barcodes (10NT, 1-24)
Discover
enables extreme multiplexing to reduce sequencing costs. Additionally, a complete automated workflow is available for labs seeking to increase throughput and minimize human error. Using cell line-derived reference standards with known mutations, we conducted a proof-of-concept study to evaluate the suitability of the NEXTFLEX Rapid XP V2 library prep kit for detecting germline mutations. These standards offer a controlled means to assess the performance of our WGS workflow, confirming its ability to identify genetic variants in sample material. This study also highlights the potential of the NEXTFLEX Rapid XP V2 kit for comprehensive genomic analysis in research applications.
Methods
Briefly, 25 ng of Mimix™ Male Genomic DNA (HD865) reference standard was used as input for whole genome library preparation. 8 replicates were made. Libraries were made using the NEXTFLEX Rapid XP V2 DNA-seq kit
 NEXTFLEX Rapid XP V2 DNA-Seq Kit for Metagenomics
Discover
and the NEXTFLEX Unique Dual Index Barcodes
NEXTFLEX Rapid XP V2 DNA-Seq Kit for Metagenomics
Discover
and the NEXTFLEX Unique Dual Index Barcodes
 NEXTFLEX Unique Dual Index Barcodes (8NT index, 1-96)
Discover
according to manufacturer’s instructions. The technicians that prepared libraries are experienced in molecular biology but had never used this library prep kit before. Once libraries were prepared, they were quantified using the Thermo Scientific® Qubit® assay, pooled and run on a S2 flow cell on Illumina® NovaSeq® 6000 platform at 2x150 bp read length. Analysis was performed using a custom script, using the VCF file provided for the Mimix reference standards and GRch38 build as reference.
NEXTFLEX Unique Dual Index Barcodes (8NT index, 1-96)
Discover
according to manufacturer’s instructions. The technicians that prepared libraries are experienced in molecular biology but had never used this library prep kit before. Once libraries were prepared, they were quantified using the Thermo Scientific® Qubit® assay, pooled and run on a S2 flow cell on Illumina® NovaSeq® 6000 platform at 2x150 bp read length. Analysis was performed using a custom script, using the VCF file provided for the Mimix reference standards and GRch38 build as reference.

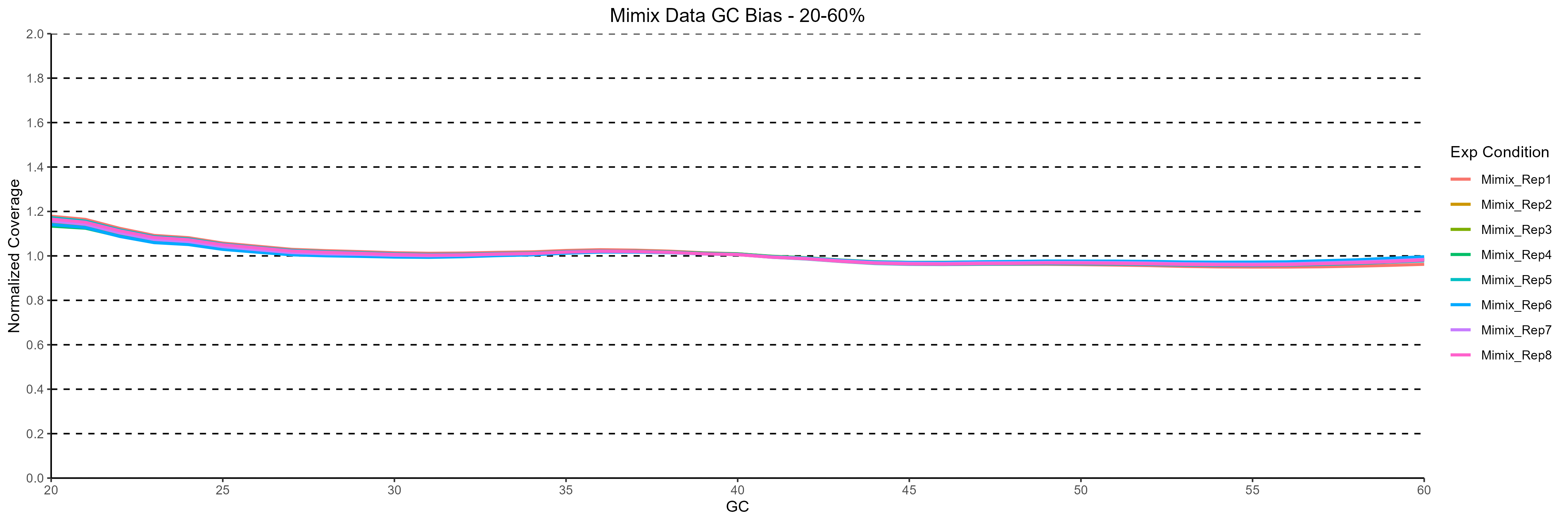
Figure 1: Normalized coverage across varying %GC content for the 8 replicates analysed. The minimal GC bias observed indicates a high degree of coverage uniformity in the data generated.
The Mimix Male Genomic DNA reference standard contains 2,663 variants with AF<10% distributed across multiple regions, including 3’UTR, introns etc. Of those, 269 are missense variants, and they are distributed across 149 genes, all of them relevant in the context of newborn screening. Missense variants are particularly significant in genome-wide screening as they result in amino acid substitutions, directly impacting protein function. For this reason, we focus our attention to them in this test. These group of variants comprises mutations classified as benign, uncertain significance and pathogenic.
Results
On average 626 M reads per sample were generated after sequencing. 98.3% aligned to human genome, corresponding to an average genome coverage of 56x. Coverage uniformity is important to ensure reliable and efficient identification of genetic variation across the genome. To assess the uniformity of coverage distribution in the data, we looked at the GC bias across the 8 replicates (Figure 1).
Concordance between expected genotype from Mimix and observed genotype was 100% for the 269 variants studied. The table below lists some of those variants indicating their ClinVar number, the reference and alternative allele, and the average coverage obtained across the 8 replicates for reach of the alleles.
Table 1: Detail of the 15 first variants analysed, all of them concordant with the expected genotype. A full list of the 269 variants can be provided upon request.
| Gene | dbSNP ID | ClinVar | Alt(%) | Ref | Alt | Ref coverage | Alt coverage |
|---|---|---|---|---|---|---|---|
| HPS1 | rs58548334 | 175335 | 69.20% | A | G | 21.8 | 47.9 |
| RBM20 | rs942077 | 53184 | 68.00% | G | C | 25.8 | 53.5 |
| BAG3 | rs3858340 | 53943 | 48.80% | C | T | 24.6 | 46.1 |
| ECHS1 | rs1049951 | 370659 | 100.00% | G | A | 0.0 | 26.9 |
| ECHS1 | rs10466126 | 1245298 | 100.00% | A | G | 0.0 | 31.3 |
| DCLRE1C | rs12768894 | 253714 | 61.80% | T | C | 19.1 | 20.1 |
| CDH23 | rs779974496 | 1481561 | 45.50% | C | T | 26.8 | 23.0 |
| CDH23 | rs1449510921 | 1319554 | 48.80% | G | A | 22.9 | 25.9 |
| CDH23 | rs1227065 | 55102 | 100.00% | A | G | 0.0 | 49.8 |
| CDH23 | rs1227051 | 55120 | 100.00% | G | A | 0.0 | 53.3 |
| CDH23 | rs41281330 | 55122 | 45.80% | G | A | 27.1 | 21.5 |
| CDH23 | rs11592462 | 55162 | 40.00% | C | G | 28.4 | 26.1 |
| BMPR1A | rs11528010 | 50221 | 72.20% | C | A | 27.1 | 21.0 |
| DBT | rs12021720 | 134332 | 100.00% | T | C | 0.0 | 51.1 |
| ATM | rs1801516 | 133907 | 31.80% | G | A | 30.3 | 30.4 |
The variants reported were inspected using IGV across the 8 replicates

Figure 2: Visualization of rs1801516, located on the exon 37 of ATM (chr 11) and leading to the change p.Asp1853Asn
Conclusion
WGS is increasingly recognized as a pivotal tool for comprehensive screening across diverse applications. Our evaluation using Mimix reference standards demonstrates that NEXTFLEX Rapid XP V2 library preparation kit delivers uniform data coverage and enhanced variant detection accuracy, even when implemented by first-time users. These results demonstrate that laboratories can confidently adopt this robust solution regardless of their prior NGS experience, supporting reliable and consistent genomic analysis with minimal technical barriers.
Featured products

NEXTFLEX Rapid XP V2 DNA-Seq Kit
- Single-tube enzymatic fragmentation, end-repair & A-tailing - DNA to sequencing-ready libraries in as little as 2.5 hours
- PCR-free option for ≥ 250 ng input plus a broad 100 pg - 1 µg range for low- and high-input samples with PCR amplification
- Built-in NEXTFLEX normalization beads - skip post-library quantitation and manual pooling
- Ultra-low adapter-dimer formation & uniform GC coverage for confident variant detection
- Up to 1,536 UDIs available for large, multiplexed studies
- 96 UDI-UMI barcode set enables molecular error-correction for rare variant calling
- Automation-ready workflow validated on Revvity Automation NGS workstations
- Quality assured through rigorous testing of every lot
The NEXTFLEX Rapid XP V2 DNA-Seq Kit unites speed, data quality, and scalability: enzymatic fragmentation streamlines the workflow, proprietary normalization beads remove an entire QC step, and an optional PCR-free path preserves library complexity. Whether processing a handful of samples or a 96-well plate on a liquid-handler, you get consistent, unbiased libraries ready for Illumina® and Element AVITI™ sequencing.

NEXTFLEX UDI Barcodes (10NT, 1-24)
- Up to 1,536 unique i5 / i7 pairs (10NT) ready to ship
- Compatible with PCR-free and PCR-amplified workflows
- ≥ 3-base Hamming distance across the entire set for robust error-correction and balanced base diversity on 2- & 4-color chemistries
- Designed to minimize index hopping that can be present with patterned flow cells.
- Compatible with Revvity library prep and TruSeq style, ligation-based workflows
- Every lot is functionally validated and tested for index purity by sequencing in an ISO 9001 facility
- Optional NEXTFLEX Universal Blockers boost on-target hybrid-capture efficiency when used with these UDIs.

NEXTFLEX Rapid XP V2 DNA-Seq Kit for Metagenomics
The NEXTFLEX Rapid XP v2 DNA-Seq Kit for Metagenomics combines fast, automation-ready library prep with built-in normalization and integrated downstream analysis. Designed for whole genome sequencing of microbial communities, purchase includes access to Cosmos-Hub®
for streamlined WGS metagenomics interpretation.
- Single-tube enzymatic fragmentation, end-repair & A-tailing. Libraries in as little as 2.5 hours
- PCR-free option for ≥250 ng input; full 100 pg–1 µg input range with amplification
- Integrated NEXTFLEX normalization beads, skip post-library quantitation and pooling
- Validated Cosmos-Hub access for taxonomic and functional WGS metagenomics analysis
- CHAMP and KEPLER pipelines support version-controlled, reproducible reporting
- Up to 1,536 UDIs available for high-throughput multiplexing
- Automation-validated on Revvity Sciclone™ and Zephyr™ NGS platforms
- Compatible with Illumina® and Element® sequencing platforms

NEXTFLEX Unique Dual Index Barcodes (8NT index, 1-96)
- UDI full length adapters for multiplexing up to 384 libraries for Illumina® and Element® sequencing platforms
- Compatible with all PCR-free and amplified TruSeq-style T-overhang library preps as drop in replacement
- Unique Dual Index design with ≥3 hamming distance reduces index hopping and sample mis-assignment on patterned flow cells
- Each lot is sequence-verified for index purity (QC by sequencing)
- Compatible with the NEXTFLEX Universal Blockers
Featured categories

Whole Genome Sequencing
At Revvity Omics, we are proud to be among the first laboratories providing whole genome sequencing as a clinical service to benefit our clients and patients.

Mimix Reference Standards
Revvity Mimix Reference Standards are all cell-line derived to maintain genomic coverage across the molecular diagnostics workflow.




























