Introduction
Within the last decade, the emergence of widely accessible methods for single-cell analysis has opened a new frontier for cell biology providing higher resolution insight into cellular heterogeneity. Single-cell technologies, known collectively as single-cell and spatial omics, enable the analysis of the genome and transcriptome of large numbers of individual cells within a sample allowing scientists to understand the nuanced impact of spatiotemporal organization of individual cells on everything from embryonic development to tumor progression and aging.
Single-cell transcriptomics enables researchers to investigate the expression profile of individual cells, uncovering the incredible heterogeneity that exists within tissues and shedding light on cell types, subtypes, and their functions. Spatial transcriptomics provides an additional layer of information, revealing the spatial context of gene expression data within a tissue or organ, and can also be used to determine the subcellular location of individual transcripts.
These techniques have the potential to revolutionize drug discovery, personalized medicine, and our ability to unravel the molecular underpinnings of various diseases, promising a future where targeted therapies and interventions are more precise and effective than ever before. To meet the needs of these advancing NGS techniques, automated sequencing platforms offer reduced costs and faster assay times, reducing months of work into days or even hours. However, while sequencing continues to improve, a new bottleneck has emerged: library preparation.
Why automate?
In this blog, we explore the issues associated with manual library preparation workflows, and the vital role of NGS automation in addressing these challenges.
The library preparation bottleneck in single-cell and spatial transcriptomics
Library preparation generally involves similar steps whether you’re analyzing DNA or RNA. Although, in the case of RNA, it’s important that the RNA is converted into complementary DNA (cDNA) before further processing. This process requires precise timing and pipetting proficiency by the operator and can take up to several hours depending on sample count.
Typically, most library preparation looks something like this:
- DNA extraction or cDNA synthesis
- DNA fragmentation
- End-repair
- A-tailing
- Adapter ligation and bead cleanup
- PCR amplification and bead cleanup
- Library quantitation
The multitude of repetitive steps not only introduces user-to-user variability but also poses a contamination risk, significantly undermining the quality and yield of your libraries. These issues can result in skewed and unreliable data, ultimately compromising the integrity of any research findings.
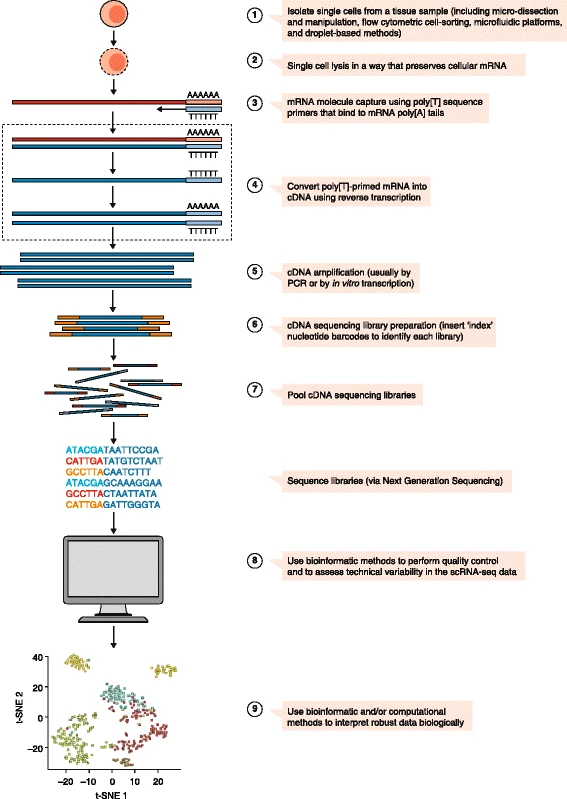
And this process is even more complicated for single-cell and spatial omics as they also require the isolation of single cells or in-situ hybridization before any DNA or RNA can be processed. While these techniques diverge in their sample processing methodologies, they share common stages such as cDNA synthesis, cDNA amplification, and PCR indexing. (Figure 1).
Source: A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications (Haque et al. Genome Medicine (2017) 9:75 - https://doi.org/10.1186/s13073-017-0467-4) Fig1.

Fig1: General workflow of single-cell RNA-sequencing (scRNA-seq) experiments. A typical scRNA-seq workflow includes most of the following steps: 1) isolation of single cells, 2) cell lysis while preserving mRNA, 3) mRNA capture, 4) reverse transcription of primed RNA into complementary DNA (cDNA), 5) cDNA amplification, 6) preparation of cDNA sequencing library, 7) pooling of sequence libraries, 8) use of bio-informatic tools to assess quality and variability, and 9) use of specialized tools to analyse and present the data. t-SNE t-distributed stochastic neighbour embedding
These steps, though crucial, are laborious, time-consuming, and highly sensitive to variation and contamination. Despite this, they are also easily standardized and automated — offering the potential to enhance reproducibility, throughput, and efficiency while reducing the labor and time required for library preparation.
Accelerating research with NGS automation
Automated library prep systems are becoming more popular, as they have the potential to overcome many of the challenges associated with manual workflows.


Image 1: NGS automation example: 10x Genomics® Chromium™ Single Cell 3' Library Kit v3 automated on Zephyr™ G3 workstation (1A Zephyr deck configuration – 1B close-up)
Selected liquid handling systems tailored to NGS library prep can now deliver a range of benefits including:
- Precision and accuracy: Automated systems execute complex protocols with exceptional precision and high reproducibility, significantly reducing the margin for error.
- Reduced risk of contamination: Human interaction with reagents and samples is a primary source of contamination in laboratory workflows. By automating the process, the risk of contamination is markedly diminished, ensuring the integrity of your experiments and the purity of your results.
- Batch-to-batch consistency: With automated systems, there's no need to fret over variations between different operators. The consistency of library preparation is maintained from one batch to the next, offering a reliable and robust starting point for scientific research.
- Increased throughput: The latest automated systems are engineered to prepare samples quickly and on a large scale with minimal to no manual interaction.
- Reduced hands-on time: Free up your scientists to focus on the science itself, rather than the mechanics of pipetting and sample preparation.
In addition, some platforms such as the Fontus™ workstation have taken an additional step by offering ready-to-go vendor-qualified methods for NGS library prep, enabling you to get started immediately rather than needing to build, optimize, and test protocols. Recent studies have demonstrated the transformative impact of automation in research. In one study, a liquid handling platform was used to automate part of a spatial transcriptomics workflow, increasing the number of samples processed per run, reducing sample preparation time, and minimizing batch effects between samples1. Using liquid handling platforms like the Fontus™ workstation, it is possible to process as many as 384 samples at one time, producing rapid and robust libraries for downstream analysis.
Another study also demonstrated the profound effect of using an automated workflow for scRNA-seq2. Using a liquid handling system, cDNA was fragmented, followed by end repair, A-tailing, and bead-based size selection. Additionally, the liquid handling system was also able to automate adapter ligation, bead cleanup and PCR indexing. Using this automated protocol, the amount of required hands-on time was reduced by more than 75%, from 4 h to 45 minutes. As such, automation allows for fast, error-free, and consistent generation of reliable libraries.
Elevate your research
As the cost and capacity of NGS continues to improve, high resolution techniques such as single cell transcriptomics and spatial transcriptomics are becoming increasingly common. However, the efficiency and throughput of library preparation techniques has not kept pace with NGS advancements, and the need for this has driven the emergence of robust, scalable, and automated library preparation protocols. Recent studies have shown the transformative impact of automation, increasing the number of samples processed, reducing preparation time, and improving the quality of libraries.
In this rapidly evolving landscape, researchers can elevate their work by embracing NGS automation. As we continue to push the boundaries of genetic research, automated library preparation represents a critical step forward, enabling faster, error-free, and reproducible generation of high-quality data. Automation is not just a convenience; it is a necessity for harnessing the full potential of NGS and advancing our understanding of the biological world.
At Revvity, we understand the critical role of NGS automation in research. Backed by 35 years of industry experience, our Fontus™ NGS liquid handler has been designed with a keen focus on user-friendliness, enhanced efficiency, and customizability to suit your needs. By automating the library preparation process our solution enables researchers to focus on the science itself and accelerate research timelines.
Discover our NGS library preparation automated methods and how you can optimize using the Fontus workstation.
References:
- Berglund, E., Saarenpää, S., Jemt, A., Gruselius, J., Larsson, L., et al. (2020). Automation of Spatial Transcriptomics library preparation to enable rapid and robust insights into spatial organization of tissues. BMC Genomics, 21, 298. https://doi.org/10.1186/s12864-020-6631-z
- Kind, D., Baskaran, P., Ramirez, F., Giner, M., Hayes, M., et al. (2022). Automation enables high-throughput and reproducible single-cell transcriptomics library preparation. SLAS Technology, 27(2), 135–142. https://doi.org/10.1016/j.slast.2021.10.018
For research use only. Not for use in diagnostic procedures.


































